plot sma
|
Contents: Description, Arguments, Usage, Examples, Images, Sub-Functions, Related Functions, Source Supported Under Version: > 1.09
Description Plot the output of sma() with the endmembers scaled to the appropriate concentration
Arguments and Return Values Arguments: the output structure of sma() and any number of options Return Value: a plot showing the measured spectrum, the modeled fit and the individual endmember concentrations represented in several ways.
Usage Syntax: plot_sma(sma_struc [, group=BOOL][, nplot=INT][, other=BOOL][, refit=INT][, round=BOOL][, stack=BOOL][, error=BOOL][, concen=BOOL][, ps=STRING][, pdf=BOOL][, min_conc=FLOAT][, long=BOOL][, i=INT][, j=INT][, x1=FLOAT][, x2=FLOAT][, y1=FLOAT][, y2=FLOAT][, lw=INT][, font_size=INT][, cm=BOOL][, non_zero=FLOAT]) sma_struc = structure returned from 'sma' Default only plots endmembers will abundances greater than .005 (0.5 percent). Modify using 'min_conc' Options: group = 1 used the 'grouped' abundances and spectra - default is the individual endmembers min_conc = minimum fraction to plot - Default is 0.005 (0.5 percent) nplot = m only plot the first m of the n endmembers found (alternative approach to min_conc other = 1 plot the sum of the rest of the endmembers found (< min_conc or > m)as 'other' round = 1 round values to nearest 5 percent. (not as sophisticated as plot_sma_groups) stack = 1 make a 'stack' plot of the endmembers refit = n rerun sma using only the first 'n' most abundance endmembers error = 1 include error values in labels diff = 1 plot the difference between the best-fit and measured spectra concen = 1 use un- normalized percentages in plot key (default is normalized to account for blackbody ps = 'basename' write to .ps file. filename is basename+i_j.ps pdf = 1 convert .ps files to .pdf (must also use 'ps' option) notitle = 1 do not add plot title using spectrum label long = 1 use long labels in key i = n only plot the ith column j = n only plot the jth row x1 = wavenumber minimum x2 = wavenumber maximum y1 = emissivity minimum y2 = emissivity maximum lw = n change line weight (default = 3) cm = 1 Plot with x-axis as Wavenumber font_size = n change font_size (default = 22) typical use: plot_sma(dqube, minc_conc=.05, other=1, stack=1, round=1) stack plot with rounded abundances. Endmembers with abundance <5% plotted as 'other' other examples: plot_sma(dqube) plot all endmembers plot_sma(dqube, refit=6, i=17) refits and plots 1st 6 endmembers for colunm 17 of qube plot_sma(dqube, error=1, ps='/myhome/figures/crater_traverse', pdf=1) plot errors and output ps and pdf files
Examples dv> b3008read($DV_EX+"/brlib_3008.hdf")
Downloading.done.
struct, 11 elements
asu_lib_id: 53x1x1 array of int, bsq format [212 bytes]
comment: Text Buffer with 53 lines of text
1: chemistry available
2: Ward's research grade sample. Band shapes and positions match well with Salisbury et al. (1992) pyrope.
3: Chemistry and XRD suggest this sample is borderline labradorite; Ab50An48Or2
4: spectrum obtained from powdered sample
5: na
6: chemistry available
7: chemistry available
8: chemistry available
9: "Sample was supplied as microcline, but chemistry and spectral character suggest microcline plus albite (perthite); Ab51An2Or47"
10: chemistry available
data: 53x1x167 array of float, bsq format [35,404 bytes]
group: Text Buffer with 53 lines of text
1: Plagioclase
2: Plagioclase
3: Plagioclase
4: Plagioclase
5: Plagioclase
6: Plagioclase
7: Clinopyroxene
8: Orthopyroxene
9: Clinopyroxene
10: Clinopyroxene
id: 53x1x1 array of int, bsq format [212 bytes]
label: Text Buffer with 53 lines of text
1: Albite WAR-5851
2: Oligoclase WAR-5804
3: Andesine BUR-240
4: Labradorite WAR-4524
5: Labradorite WAR-RGAND01
6: Anorthite BUR-340
7: Hedenbergite manganoan DSM-HED01
8: Bronzite NMNH-93527
9: Diopside WAR-5780
10: Augite HS-119.4B
owner: Text Buffer with 53 lines of text
1: ASU
2: ASU
3: ASU
4: ASU
5: ASU
6: ASU
7: ASU
8: ASU
9: ASU
10: ASU
shortlabel: Text Buffer with 53 lines of text
1: Albite
2: Oligoclase
3: Andesine
4: Labradorite
5: Labradorite
6: Anorthite
7: Hedenbergite
8: Bronzite
9: Diopside
10: Augite
sizerange: Text Buffer with 53 lines of text
1: 710-1000 microns
2: 710-1000 microns
3: 710-1000 microns
4: 710-1000 microns
5: 710-1000 microns
6: 710-1000 microns
7: 710-1000 microns
8: 710-1000 microns
9: 710-1000 microns
10: 250-1000 microns
source: Text Buffer with 53 lines of text
1: "Bird Creek, Ontario Canada"
2: "Navajo Reservation, Ant Hill, Arizona"
3: "Big Tujunga Canyon, L.A. Co. CA"
4: "Cusihuiriachic, San Juanito, Mexico"
5: Ward's
6: "Grass Valley, CA"
7: "Iron Cap Prospect, AZ"
8: "Paul's Island, Labrador, Canada"
9: "El Paso County, CO"
10: "Oaxaca, Mexico"
xaxis: 1x1x167 array of float, bsq format [668 bytes]
dv> a1 = read($DV_EX+"/a1_mini_tes_spectrum.vic")
Downloading.done.
.1x1x167 array of double, bip format [1,336 bytes]
dv> s1 = sma(a1, b3008, sort=1, group=1, exclude=37//38//40, notchco2=1, wave1=380, wave2=1280)
struct, 23 elements
notchco2: struct, 6 elements
wco2: 74.05999756
co2chanlow: 27
co2chanhigh: 41
dwavex: 9.984985352
co2low: 594.9400024
co2high: 743.0599976
xaxis: 1x1x154 array of float, bsq format [616 bytes]
forcedlib: struct, 1 elements
label: "none"
excluded: 3x1x1 array of int, bsq format [12 bytes]
covm: 13x13x1 array of double, bsq format [1,352 bytes]
bb: 0.3020044267
bberror: 0.01848441921
normconc: 1x1x53 array of float, bsq format [212 bytes]
sort: 1x1x50 array of int, bsq format [200 bytes]
sort_full: 1x1x53 array of int, bsq format [212 bytes]
measured: 1x1x154 array of float, bip format [616 bytes]
modeled: 1x1x154 array of float, bsq format [616 bytes]
conc: 1x1x53 array of float, bsq format [212 bytes]
rms: 0.001185287023
nsamples: 77
endlib: struct, 11 elements
asu_lib_id: 53x1x1 array of int, bsq format [212 bytes]
comment: Text Buffer with 53 lines of text
1: chemistry available
2: Ward's research grade sample. Band shapes and positions match well with Salisbury et al. (1992) pyrope.
3: Chemistry and XRD suggest this sample is borderline labradorite; Ab50An48Or2
4: spectrum obtained from powdered sample
5: na
6: chemistry available
7: chemistry available
8: chemistry available
9: "Sample was supplied as microcline, but chemistry and spectral character suggest microcline plus albite (perthite); Ab51An2Or47"
10: chemistry available
data: 53x1x154 array of float, bsq format [32,648 bytes]
group: Text Buffer with 53 lines of text
1: Plagioclase
2: Plagioclase
3: Plagioclase
4: Plagioclase
5: Plagioclase
6: Plagioclase
7: Clinopyroxene
8: Orthopyroxene
9: Clinopyroxene
10: Clinopyroxene
id: 53x1x1 array of int, bsq format [212 bytes]
label: Text Buffer with 53 lines of text
1: Albite WAR-5851
2: Oligoclase WAR-5804
3: Andesine BUR-240
4: Labradorite WAR-4524
5: Labradorite WAR-RGAND01
6: Anorthite BUR-340
7: Hedenbergite manganoan DSM-HED01
8: Bronzite NMNH-93527
9: Diopside WAR-5780
10: Augite HS-119.4B
owner: Text Buffer with 53 lines of text
1: ASU
2: ASU
3: ASU
4: ASU
5: ASU
6: ASU
7: ASU
8: ASU
9: ASU
10: ASU
shortlabel: Text Buffer with 53 lines of text
1: Albite
2: Oligoclase
3: Andesine
4: Labradorite
5: Labradorite
6: Anorthite
7: Hedenbergite
8: Bronzite
9: Diopside
10: Augite
sizerange: Text Buffer with 53 lines of text
1: 710-1000 microns
2: 710-1000 microns
3: 710-1000 microns
4: 710-1000 microns
5: 710-1000 microns
6: 710-1000 microns
7: 710-1000 microns
8: 710-1000 microns
9: 710-1000 microns
10: 250-1000 microns
source: Text Buffer with 53 lines of text
1: "Bird Creek, Ontario Canada"
2: "Navajo Reservation, Ant Hill, Arizona"
3: "Big Tujunga Canyon, L.A. Co. CA"
4: "Cusihuiriachic, San Juanito, Mexico"
5: Ward's
6: "Grass Valley, CA"
7: "Iron Cap Prospect, AZ"
8: "Paul's Island, Labrador, Canada"
9: "El Paso County, CO"
10: "Oaxaca, Mexico"
xaxis: 1x1x154 array of float, bsq format [616 bytes]
error: 1x1x53 array of float, bsq format [212 bytes]
spectral_range: "Channels 6-82"
spectral_channel_low: 6
spectral_channel_high: 82
wave1: 380
wave2: 1280
grouped: struct, 7 elements
grouped_conc: 1x1x13 array of float, bsq format [52 bytes]
grouped_error: 1x1x13 array of float, bsq format [52 bytes]
grouped_labels: Text Buffer with 13 lines of text
1: Plagioclase
2: Clinopyroxene
3: Orthopyroxene
4: Olivine
5: Pigeonite
6: Mars Dust
7: Hematite
8: Pyroxenoid
9: Mars Surface
10: Oxide
grouped_bb: 0.3020044267
grouped_bberror: 0.01854504272
grouped_sort_full: 1x1x13 array of byte, bsq format [13 bytes]
grouped_normconc: 1x1x13 array of float, bsq format [52 bytes]
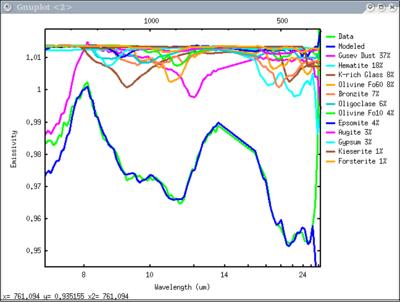
dv> plot_sma(s1)
doing i = 1 j = 1
|
DavinciWiki Mini-Nav Bar Contents
Contact Developers
All other topics
Related Functions
Recent Library Changes Created On: 08-20-2010 |